U-net for solving PDEs
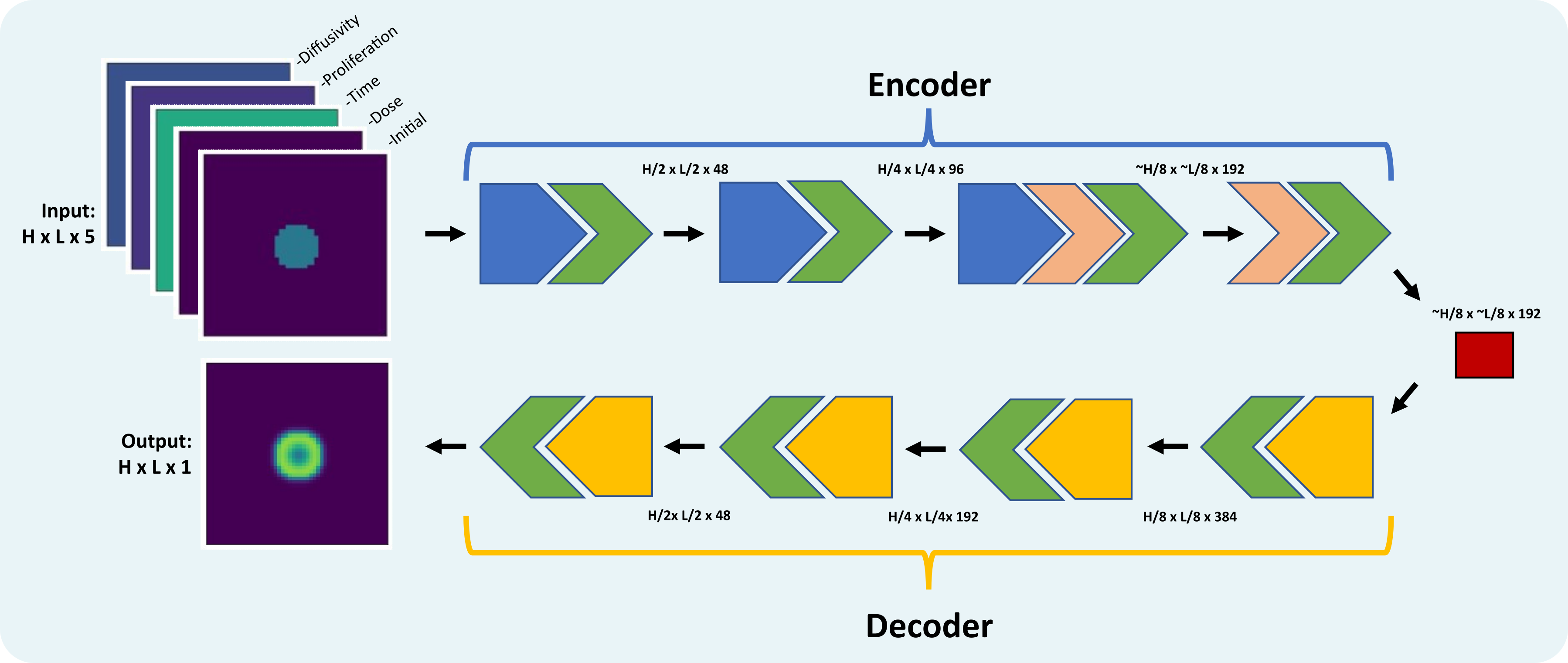
This project served as an introduction to surrogate models using convolutional neural networks. Working the PDEs found in my dissertation work, I built and trained a CNN with a U-net architecture to replace the FDM numerical solver for the reaction diffusion equation. This model was trained and evaluated on in silico data based on parameter ranges used for fitting patient data.
The encoder takes 2D feature map inputs describing the initial condition, growth parameters, time for simulation and doses from radiation. The decoder takes the latent vector and outputs the simulation at the time described by the input. The inputs for training and testing were rather simple, consisting of only radial growth, but was very successful with only a small number of samples in a training set.
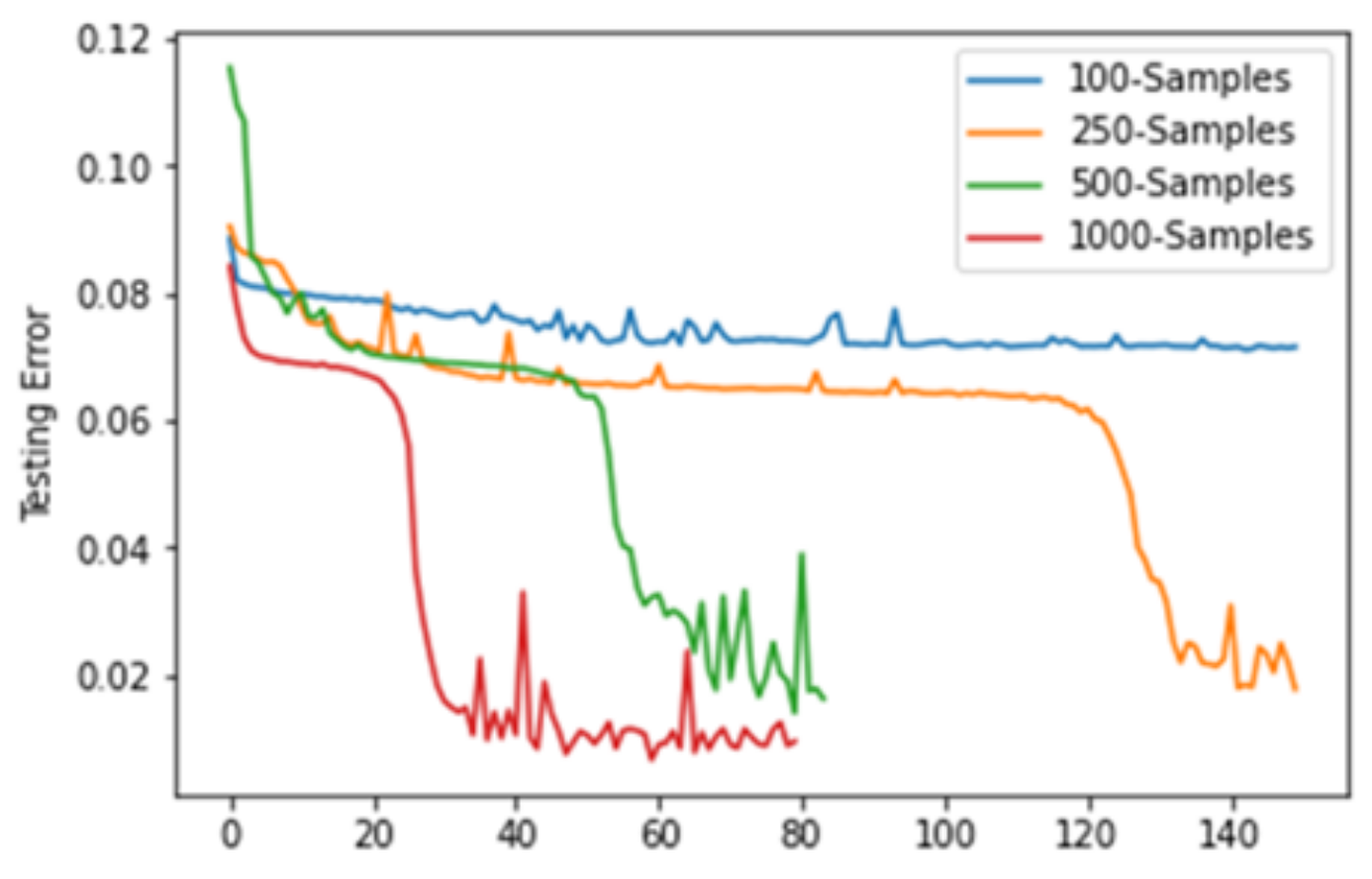
This work played a large part in my interest in computational reductions, and led me into the dimensionality reduction found here.